# 関数
library(dplyr)
library(ggplot2)
seed <- 20230430
location_x <- location_y <- 50
fun_sim <- function(n_x, n_y, location_x, location_y, scale_x, scale_y) {
x <- rlogis(n = n_x, location = location_x, scale = scale_x)
y <- rlogis(n = n_y, location = location_y, scale = scale_y)
value <- c(x, y)
key <- c(rep("x", n_x), rep("y", n_y))
df <- data.frame(key, value)
g <- ggplot(data = df, mapping = aes(x = value, fill = key)) + geom_histogram(col = "white", position = "identity", alpha = 0.7) + theme_minimal() + theme(legend.title = element_blank())
result_ansari <- ansari.test(x, y, alternative = "two.sided")
return(list(g = g, result_ansari = result_ansari, result_median = list(x = median(x), y = median(y))))
}
# 以降有意水準は5%とする。Rで統計的仮説検定:アンサリ・ブラッドリー検定
Rでデータサイエンス
アンサリ・ブラッドリー検定
- 中央値の等しい2つの群の分散(scale)同一性の検定
- ノンパラメトリック検定(正規分布に従わない群に利用できる)
- 帰無仮説:2つの群の分散(scale)は等しい
サンプル
set.seed(seed = seed)
# サンプルサイズは同一
n_x <- 100
n_y <- 100
# 分散は同一
scale_x <- 3
scale_y <- 3
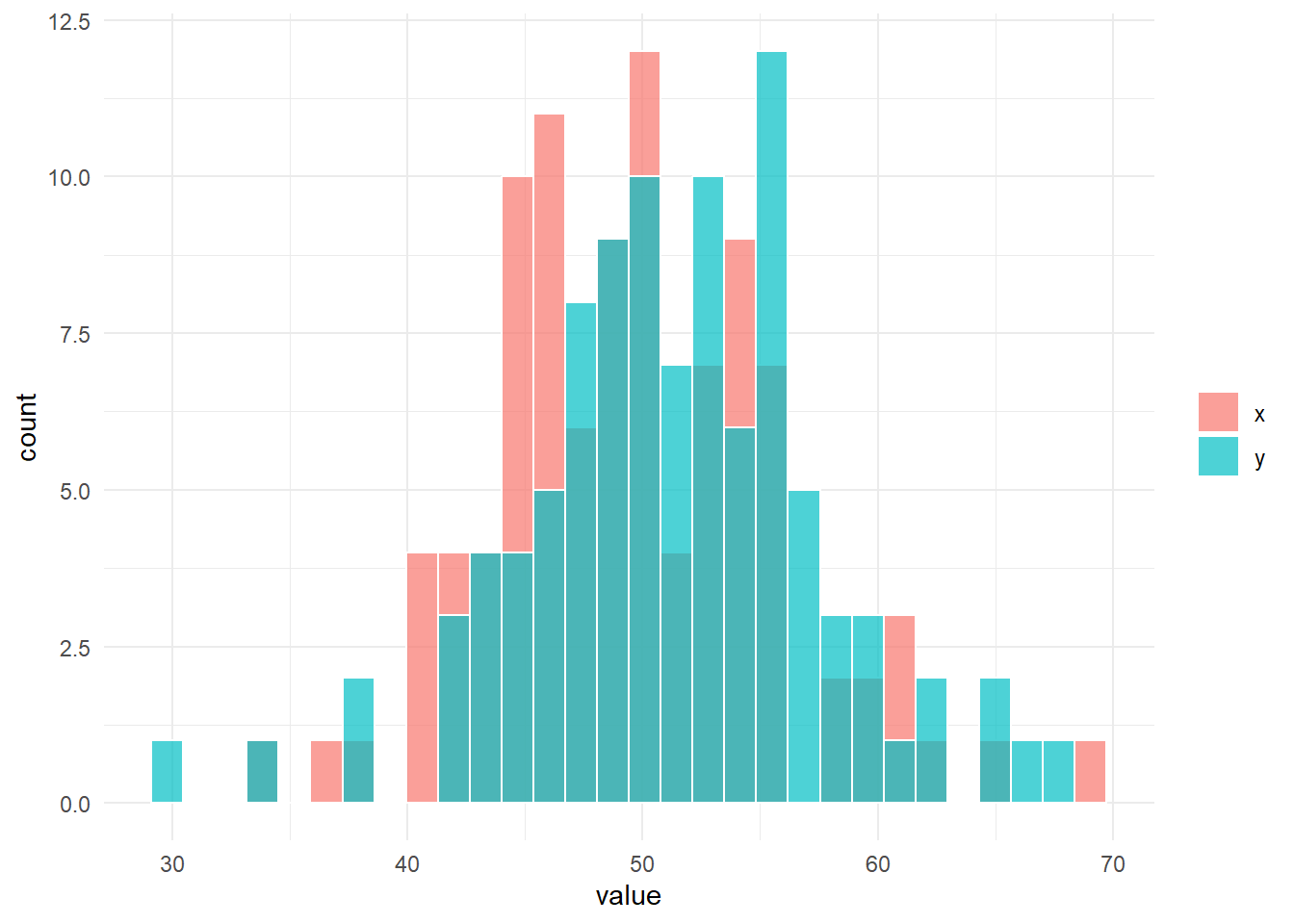
fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
# 帰無仮説は棄却されない。$g
$result_ansari
Ansari-Bradley test
data: x and y
AB = 5047, p-value = 0.9883
alternative hypothesis: true ratio of scales is not equal to 1
$result_median
$result_median$x
[1] 49.07661
$result_median$y
[1] 51.37151set.seed(seed = seed)
# サンプルサイズは同一
n_x <- 100
n_y <- 100
# 分散は異なる
scale_x <- 3
scale_y <- 5
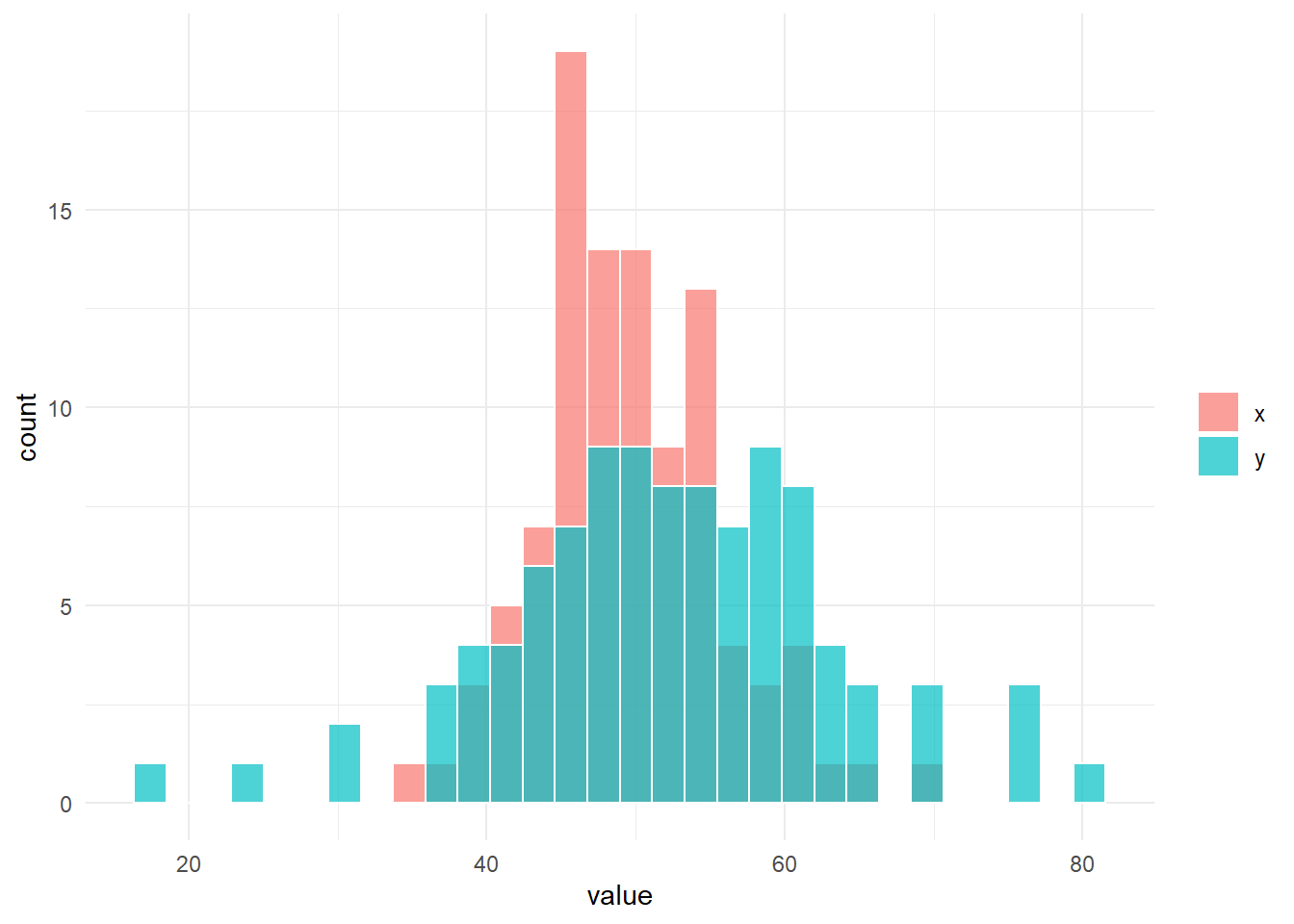
fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
# 帰無仮説は棄却される。$g
$result_ansari
Ansari-Bradley test
data: x and y
AB = 5749, p-value = 0.0006355
alternative hypothesis: true ratio of scales is not equal to 1
$result_median
$result_median$x
[1] 49.07661
$result_median$y
[1] 52.28586set.seed(seed = seed)
# サンプルサイズは異なる
n_x <- 100
n_y <- 200
# 分散は同一
scale_x <- 3
scale_y <- 3
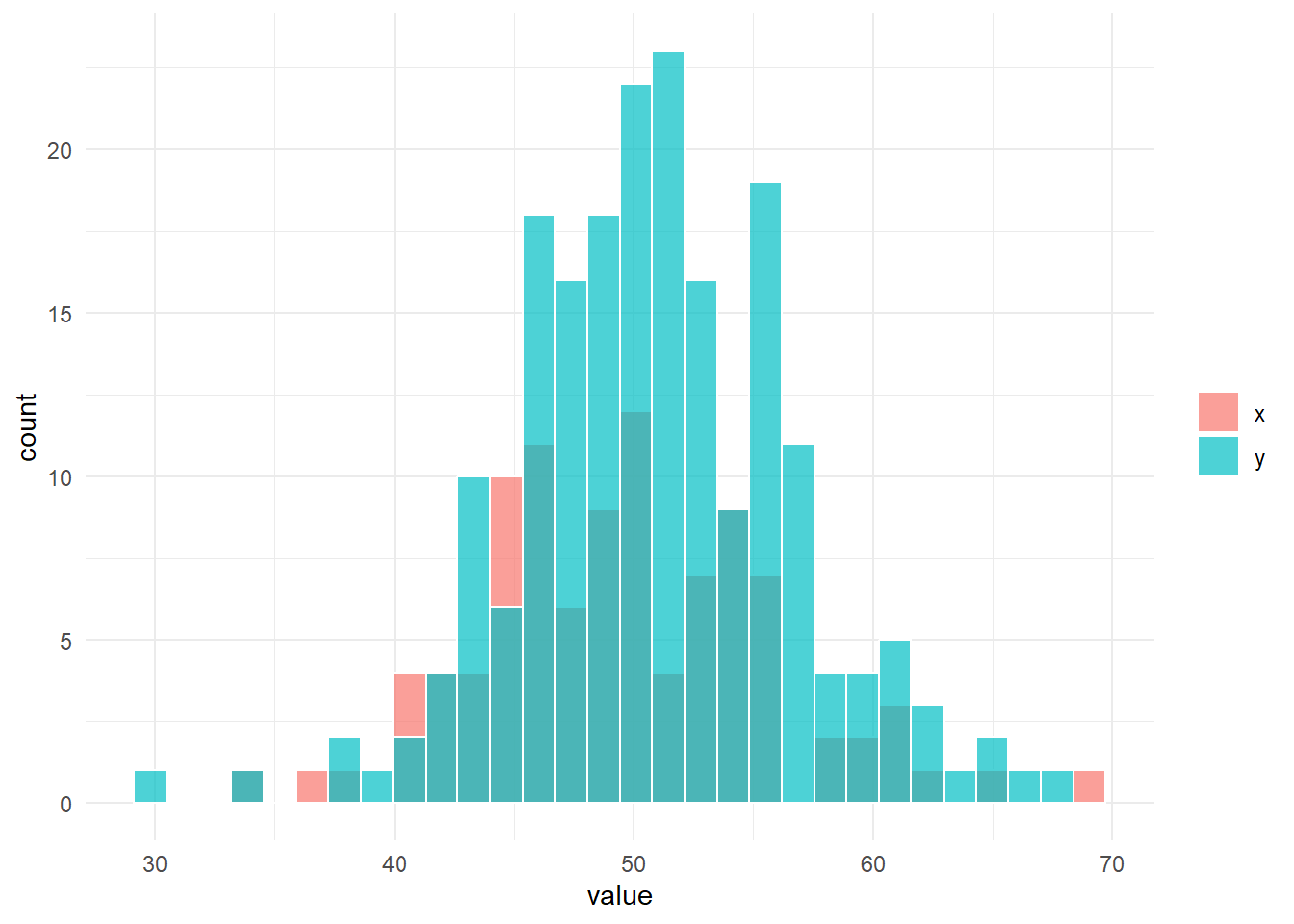
fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
# 帰無仮説は棄却されない。$g
$result_ansari
Ansari-Bradley test
data: x and y
AB = 7194, p-value = 0.3148
alternative hypothesis: true ratio of scales is not equal to 1
$result_median
$result_median$x
[1] 49.07661
$result_median$y
[1] 50.73988set.seed(seed = seed)
# サンプルサイズは異なる
n_x <- 100
n_y <- 200
# 分散は異なる
scale_x <- 3
scale_y <- 5
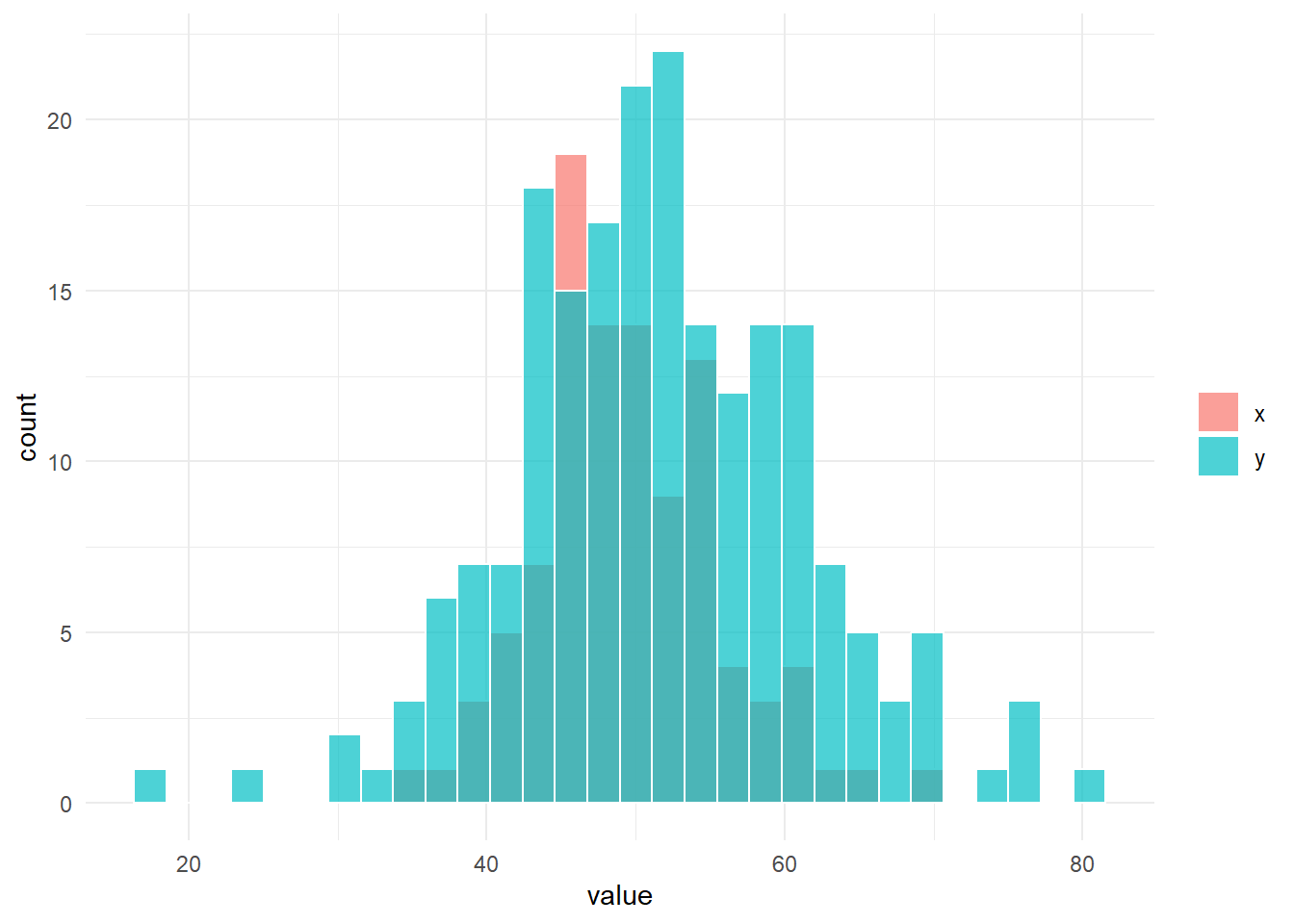
fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
# 帰無仮説は棄却される。$g
$result_ansari
Ansari-Bradley test
data: x and y
AB = 8639, p-value = 0.002104
alternative hypothesis: true ratio of scales is not equal to 1
$result_median
$result_median$x
[1] 49.07661
$result_median$y
[1] 51.23313シミュレーション
set.seed(seed = seed)
iter <- 1000
result_ansari_p <- vector()
# サンプルサイズは同一
n_x <- 100
n_y <- 100
# 分散は同一
scale_x <- 3
scale_y <- 3
for (iii in seq(iter)) {
result <- fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
result_ansari_p[iii] <- result$result_ansari$p.value
}
sum(result_ansari_p < 0.05)/iter[1] 0.053set.seed(seed = seed)
iter <- 1000
result_ansari_p <- vector()
# サンプルサイズは同一
n_x <- 100
n_y <- 100
# 分散は異なる
scale_x <- 3
scale_y <- 5
for (iii in seq(iter)) {
result <- fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
result_ansari_p[iii] <- result$result_ansari$p.value
}
1 - sum(result_ansari_p < 0.05)/iter[1] 0.061set.seed(seed = seed)
iter <- 1000
result_ansari_p <- vector()
# サンプルサイズは異なる
n_x <- 100
n_y <- 200
# 分散は同一
scale_x <- 3
scale_y <- 3
for (iii in seq(iter)) {
result <- fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
result_ansari_p[iii] <- result$result_ansari$p.value
}
sum(result_ansari_p < 0.05)/iter[1] 0.056set.seed(seed = seed)
iter <- 1000
result_ansari_p <- vector()
# サンプルサイズは異なる
n_x <- 100
n_y <- 200
# 分散は異なる
scale_x <- 3
scale_y <- 5
for (iii in seq(iter)) {
result <- fun_sim(n_x = n_x, n_y = n_y, location_x = location_x, location_y = location_y, scale_x = scale_x, scale_y = scale_y)
result_ansari_p[iii] <- result$result_ansari$p.value
}
1 - sum(result_ansari_p < 0.05)/iter[1] 0.015コード
# 関数コード
methods("ansari.test")[1] ansari.test.default* ansari.test.formula*
see '?methods' for accessing help and source codegetS3method(f = "ansari.test", class = "default")function (x, y, alternative = c("two.sided", "less", "greater"),
exact = NULL, conf.int = FALSE, conf.level = 0.95, ...)
{
alternative <- match.arg(alternative)
if (conf.int) {
if (!((length(conf.level) == 1L) && is.finite(conf.level) &&
(conf.level > 0) && (conf.level < 1)))
stop("'conf.level' must be a single number between 0 and 1")
}
DNAME <- paste(deparse1(substitute(x)), "and", deparse1(substitute(y)))
x <- x[complete.cases(x)]
y <- y[complete.cases(y)]
m <- as.integer(length(x))
if (is.na(m) || m < 1L)
stop("not enough 'x' observations")
n <- as.integer(length(y))
if (is.na(n) || n < 1L)
stop("not enough 'y' observations")
N <- m + n
r <- rank(c(x, y))
STATISTIC <- sum(pmin(r, N - r + 1)[seq_along(x)])
TIES <- (length(r) != length(unique(r)))
if (is.null(exact))
exact <- ((m < 50L) && (n < 50L))
if (exact && !TIES) {
pansari <- function(q, m, n) .Call(C_pAnsari, q, m, n)
PVAL <- switch(alternative, two.sided = {
if (STATISTIC > ((m + 1)^2%/%4 + ((m * n)%/%2)/2)) p <- 1 -
pansari(STATISTIC - 1, m, n) else p <- pansari(STATISTIC,
m, n)
min(2 * p, 1)
}, less = 1 - pansari(STATISTIC - 1, m, n), greater = pansari(STATISTIC,
m, n))
if (conf.int) {
qansari <- function(p, m, n) .Call(C_qAnsari, p,
m, n)
alpha <- 1 - conf.level
x <- sort(x)
y <- sort(y)
ab <- function(sig) {
rab <- rank(c(x/sig, y))
sum(pmin(rab, N - rab + 1)[seq_along(x)])
}
ratio <- outer(x, y, `/`)
aratio <- ratio[ratio >= 0]
sigma <- sort(aratio)
cci <- function(alpha) {
u <- absigma - qansari(alpha/2, m, n)
l <- absigma - qansari(1 - alpha/2, m, n)
if (length(u[u >= 0]) == 0L || length(l[l > 0]) ==
0L) {
warning("samples differ in location: cannot compute confidence set, returning NA")
return(c(NA, NA))
}
u[u < 0] <- NA
uci <- min(sigma[which(u == min(u, na.rm = TRUE))])
l[l <= 0] <- NA
lci <- max(sigma[which(l == min(l, na.rm = TRUE))])
if (uci > lci) {
l <- absigma - qansari(alpha/2, m, n)
u <- absigma - qansari(1 - alpha/2, m, n)
u[u < 0] <- NA
uci <- min(sigma[which(u == min(u, na.rm = TRUE))])
l[l <= 0] <- NA
lci <- max(sigma[which(l == min(l, na.rm = TRUE))])
}
c(uci, lci)
}
cint <- if (length(sigma) < 1L) {
warning("cannot compute confidence set, returning NA")
c(NA, NA)
}
else {
absigma <- sapply(sigma + c(diff(sigma)/2, sigma[length(sigma)] *
1.01), ab)
switch(alternative, two.sided = cci(alpha), greater = c(cci(alpha *
2)[1L], Inf), less = c(0, cci(alpha * 2)[2L]))
}
attr(cint, "conf.level") <- conf.level
u <- absigma - qansari(0.5, m, n)
sgr <- if (length(sgr <- sigma[u <= 0]) == 0L)
NA
else max(sgr)
sle <- if (length(sle <- sigma[u > 0]) == 0L)
NA
else min(sle)
ESTIMATE <- mean(c(sle, sgr))
}
}
else {
EVEN <- ((N%%2L) == 0L)
normalize <- function(s, r, TIES, m = length(x), n = length(y)) {
z <- if (EVEN)
s - m * (N + 2)/4
else s - m * (N + 1)^2/(4 * N)
if (!TIES) {
SIGMA <- if (EVEN)
sqrt((m * n * (N + 2) * (N - 2))/(48 * (N -
1)))
else sqrt((m * n * (N + 1) * (3 + N^2))/(48 *
N^2))
}
else {
r <- rle(sort(pmin(r, N - r + 1)))
SIGMA <- if (EVEN)
sqrt(m * n * (16 * sum(r$lengths * r$values^2) -
N * (N + 2)^2)/(16 * N * (N - 1)))
else sqrt(m * n * (16 * N * sum(r$lengths * r$values^2) -
(N + 1)^4)/(16 * N^2 * (N - 1)))
}
z/SIGMA
}
p <- pnorm(normalize(STATISTIC, r, TIES))
PVAL <- switch(alternative, two.sided = 2 * min(p, 1 -
p), less = 1 - p, greater = p)
if (conf.int && !exact) {
alpha <- 1 - conf.level
ab2 <- function(sig, zq) {
r <- rank(c(x/sig, y))
s <- sum(pmin(r, N - r + 1)[seq_along(x)])
TIES <- (length(r) != length(unique(r)))
normalize(s, r, TIES, length(x), length(y)) -
zq
}
srangepos <- NULL
srangeneg <- NULL
if (length(x[x > 0]) && length(y[y > 0]))
srangepos <- c(min(x[x > 0], na.rm = TRUE)/max(y[y >
0], na.rm = TRUE), max(x[x > 0], na.rm = TRUE)/min(y[y >
0], na.rm = TRUE))
if (length(x[x <= 0]) && length(y[y < 0]))
srangeneg <- c(min(x[x <= 0], na.rm = TRUE)/max(y[y <
0], na.rm = TRUE), max(x[x <= 0], na.rm = TRUE)/min(y[y <
0], na.rm = TRUE))
if (any(is.infinite(c(srangepos, srangeneg)))) {
warning("cannot compute asymptotic confidence set or estimator")
conf.int <- FALSE
}
else {
ccia <- function(alpha) {
statu <- ab2(srange[1L], zq = qnorm(alpha/2))
statl <- ab2(srange[2L], zq = qnorm(alpha/2,
lower.tail = FALSE))
if (statu > 0 || statl < 0) {
warning("samples differ in location: cannot compute confidence set, returning NA")
return(c(NA, NA))
}
u <- uniroot(ab2, srange, tol = 0.0001, zq = qnorm(alpha/2))$root
l <- uniroot(ab2, srange, tol = 0.0001, zq = qnorm(alpha/2,
lower.tail = FALSE))$root
sort(c(u, l))
}
srange <- range(c(srangepos, srangeneg), na.rm = FALSE)
cint <- switch(alternative, two.sided = ccia(alpha),
greater = c(ccia(alpha * 2)[1L], Inf), less = c(0,
ccia(alpha * 2)[2L]))
attr(cint, "conf.level") <- conf.level
statu <- ab2(srange[1L], zq = 0)
statl <- ab2(srange[2L], zq = 0)
if (statu > 0 || statl < 0) {
ESTIMATE <- NA
warning("cannot compute estimate, returning NA")
}
else ESTIMATE <- uniroot(ab2, srange, tol = 0.0001,
zq = 0)$root
}
}
if (exact && TIES) {
warning("cannot compute exact p-value with ties")
if (conf.int)
warning("cannot compute exact confidence intervals with ties")
}
}
names(STATISTIC) <- "AB"
RVAL <- list(statistic = STATISTIC, p.value = PVAL, null.value = c(`ratio of scales` = 1),
alternative = alternative, method = "Ansari-Bradley test",
data.name = DNAME)
if (conf.int)
RVAL <- c(RVAL, list(conf.int = cint, estimate = c(`ratio of scales` = ESTIMATE)))
class(RVAL) <- "htest"
return(RVAL)
}
<bytecode: 0x000001e386b4ef58>
<environment: namespace:stats>参考引用資料
最終更新
Sys.time()[1] "2024-04-04 04:00:48 JST"R、Quarto、Package
R.Version()$version.string[1] "R version 4.3.3 (2024-02-29 ucrt)"quarto::quarto_version()[1] '1.4.542'packageVersion(pkg = "tidyverse")[1] '2.0.0'